Introduction
This page is draft material for a paper on lightweight data systems: Something you build in eight hours on the cloud that elevates your data from ‘a mess’ to ‘a usable mess’… or still better ‘a usable organized mess’ or dare we hope ‘a usable organized mess that makes some sense’.
Our toolbox includes
- Existing data systems from AralDIF to NASA DAAC
- Web frameworks (Django, Flask, Bottle, …)
- Code repos (GitHub, GitLab, …)
- Jupyter notebooks
- The public cloud
- Specifically cloud-hosted services that are free or cheap
- Specifically cloud storage that is cheap
Links
Some organizing ideas
Abstract?
We describe here a process of creating a data system within a research program which is implicitly about sharing, collaboration, reproducibility, and accelerated pace in data-driven science. This thesis is part of a shift in emphasis that borrows heavily from both commercial and open software development paradigms that we trace to one central idea: The lifespan and value of data are not intrinsically known at the outset (owing to the very nature of research) but in fact are commonly undervalued in traditional approaches to data curation. There are several instructive contrasts between success criteria in research software development versus commercial software development versus open software development. Here we borrow from these contrasts to illustrate the cultural and technical shift necessary to accomplish the normative assertion that the value of data can be better realized through the cultural shift of changing practices. The research program has traditionally had as its end goal a peer-reviewed publication and this has contributed to the de facto conservative life expectancy of source data. The following postulates are the basis for the lightweight data system framework presented here:
- Data-driven research can be roughly divided into exploratory and perfunctory tasks
- Open source solutions are built by software developers to solve common problems in a robust manner
- Open source solutions have lifespans related to their degree of success and usefulness
- Commercial software development is rigorous owing to the cost of failure
- Commercial software development is commonly managed by non-developers who perform important supporting tasks
- The public cloud is globally available as a powerful low-cost computing platform
- Each research domain has its own idiosyncratic formats, practices and tools
- Within and often across research domains – despite idiosyncracies – common objectives emerge in service to both perfunctory and exploratory tasks
- Time is a research group’s primary and most valuable commodity; followed by qualified people; followed by funding
- The Scutero principle: Given the primacy of time: Never build any more than is absolutely necessary
- Corollary: Work from design patterns
- Corollary: Be prepared to invest time learning what ‘minimum necessary’ actually is
- Corollary: Design patterns are one step from cookbook recipes; which can reduce development time
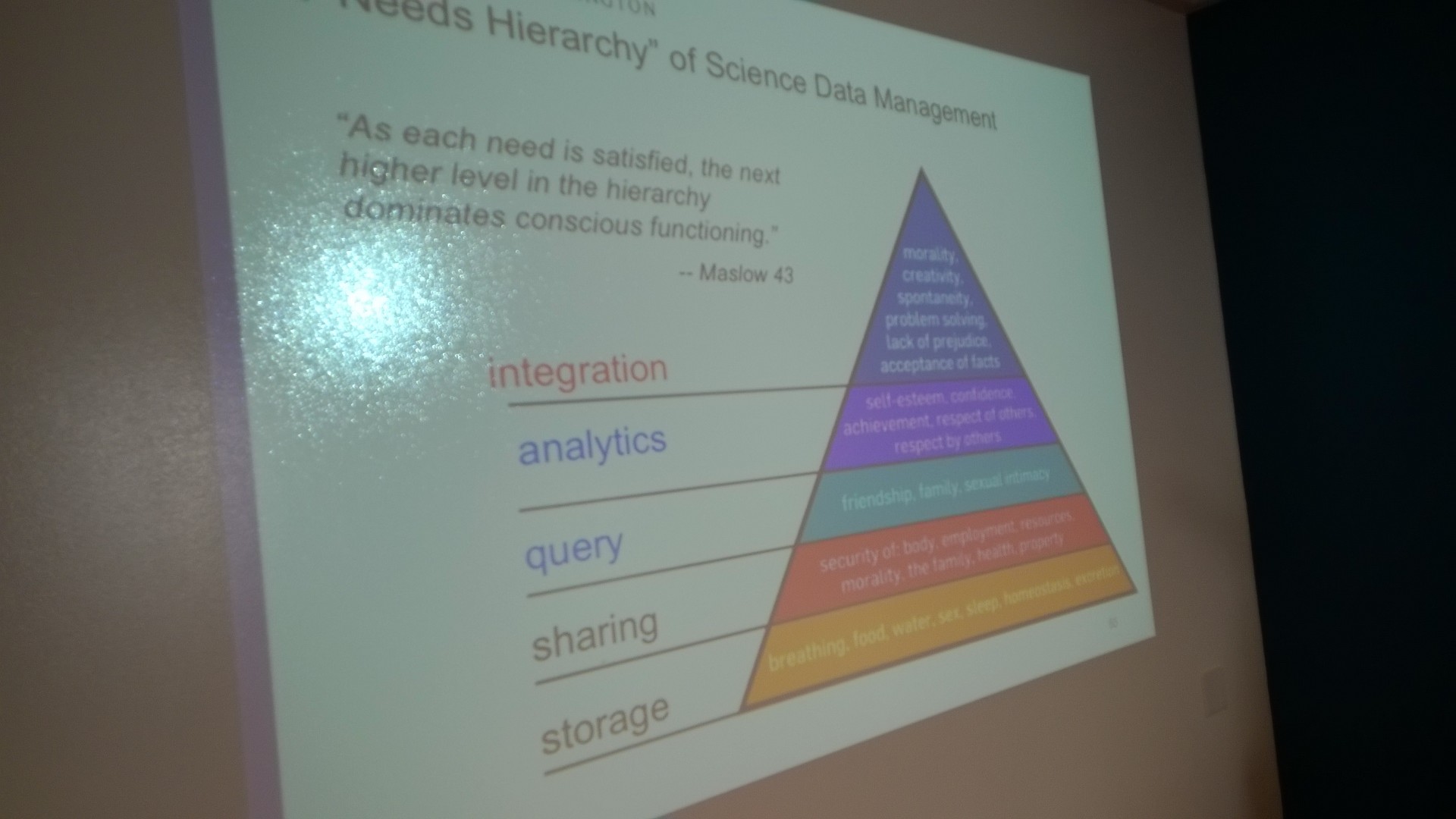
- We Howe principle: The Pyramid of human stability has a data-driven research analog which we develop and invert as follows
- Base: Storage
- Upon the base: Sharing data
- Upon sharing: Query
- Upon query: Analytics
- Upon analytics: Integration
- Inversion: Integration from the outset is a project management strategy
- Integration means making the connectivity between various moving parts of a project work together
- The concept arose over countless failures where these interfaces generated friction
- Integration amounts to devoting careful thought to what the end result looks like, how it functions
- Integration often involves two important human considerations: Use cases and stakeholders
- The positive admonition would be ‘Ignore integration at your peril’
- Integration means making the connectivity between various moving parts of a project work together
- Execution as artifact (not part of the original meeting)
- We can now create Docker images, AMIs, Kubernetes etc etc that preserve execution environments with data
- The Lightweight version of this is some form of Jupyter but it can be extended to heavy lifting
- It would be good to indicate a cost order of magnitude for this as well
- Any domain can generate a data to technology mapping that describes building a LWDS
- Our emphasis here has been on geosciences
- We might expand this paper to include genomics and something like neuroimagng
- Regardless: We claim to cover only a fraction of the domain detail
- With the idea that ‘well begun is half done’
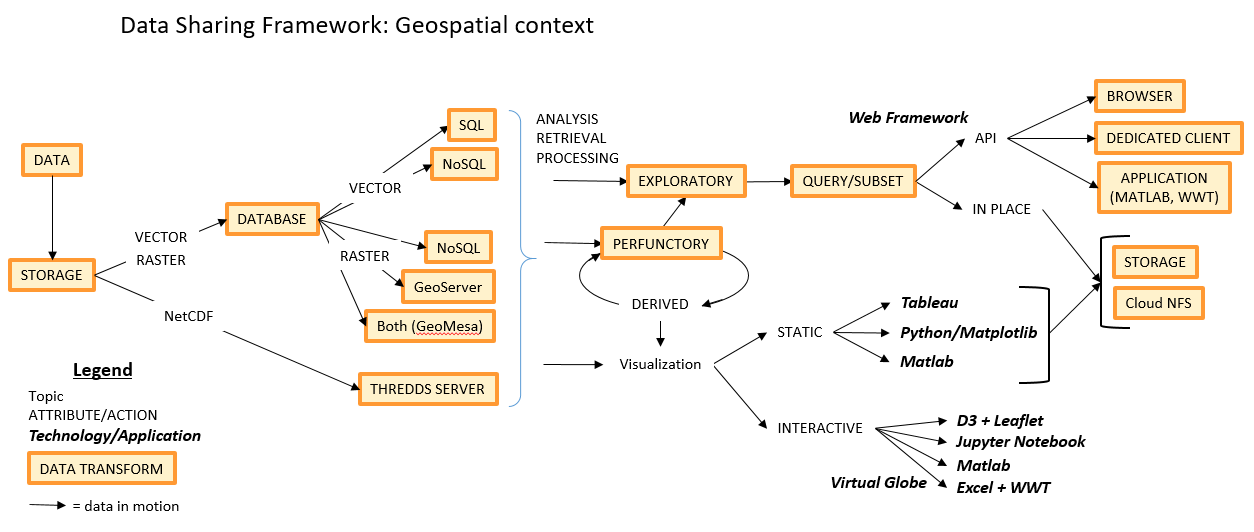
Geospatial domain
Genomics domain
Researcher remarks
- GCP as interesting alternative to AWS
- particularly impressed by Google Genomics
- ISB is a great resource for supporting end-user efforts. (see kilroy email thread for details)
-
- on Google Cloud console and cost estimator tool
- Delta? Big Query ~ Big Table
- Iinitial efforts led to a very expensive outer join in Big Query > free tier
- (which was good: it was a very sub-optimal query
- Based on conversation with Google solution architect:
- Big Table requires about $1000/month of infrastructure
- ? not a clear on-ramp for testing it out at a small scale before ramping up
- particularly for someone with limited experience with NOSQL
- Iinitial efforts led to a very expensive outer join in Big Query > free tier
- particularly impressed by Google Genomics
Research project
- TOPMed project
- Aggregating whole genome sequence data from many people across multiple studies
- Goal: Use this aggregated data to enable statistical analysis to search for associations between genetic variants and phenotypes (including common diseases) that arise from complex genetic interactions.
- When an individual’s genome is sequenced new variants are observed and added to a list of “variants we’ve observed in TOPMed”.
- We currently have about 200e6 such variants; <500e6+>
- With collaborator XL: Annotate each of those variants with about 1,000 descriptors.
- XL has built a tool called WGSA
- Includes annotation for all the possible 8,584,031,106 human single nucleotide variants.
- WGSA uses the Hg19 reference build of the human genome
- 2,897,310,462 annotated bases details
- 2,897,310,462 * 3 = 8,691,931,386.
- Looks like Hg19 includes 239,850,802 N bases, which are “unknown bases in the assembly marked by N”.
- Includes annotation for all the possible 8,584,031,106 human single nucleotide variants.
- The first expensive operation is intersect our 200e6 variants with the XL list of 8,584,031,106 potential variants
- Retrieve a set of annotation for the TOPMed variants.
- Currently, XL does this on AWS or his local infrastructure
- days to weeks
- Ref conversation with SM and Scripps about incorporating WGSA resource into the myvariant.info webapp (also on AWS)
- Backed by a mongodb and elasticsearch with a scaleable python/tornado front end
- Next expensive operation would be to aggregate variants from the TOPMed-specific list by filtering WGSA results by field.
- For example: Define a set of about 40,000 genomic ranges that define genes.
- I might want a list of all the variants in TOPMed studies that fall within each of those 40,000 ranges.
- Currently WGSA annotation results from XL are a set of large .gz tab-delim text files (one per chromosome).
- Parse these files and import them into an unoptimized mysql database > do a join as described above, prohibitively slow
- The import takes on the order of a week per chromosome (could be parallelized)
- Using a stored procedure can get the variants-by-gene list for a single chromosome in about 20 minutes.
- Parse these files and import them into an unoptimized mysql database > do a join as described above, prohibitively slow
- This approach is currently plagued with perf bottlenecks and will not scale any further
- XL has built a tool called WGSA
- Ideal path: Build an infrastructure like myvariant.info that will currently require
- parsing the text to JSON
- import to a NOSQL document store
- index it
- perform queries on it
From the photos… some text
- Creating a data resource for 2 groups is equivalent to creating a data resource for 10,000 groups (on the cloud)
- Terms to review and incorporate include
- Scale
- Micro-LWDS?
- Small: SQLShare
- Small: Geotraces
- Large: Heidi Sosik
- Large: Ameriflux
- National: NASA DAACs
- Meta: Earthcube
- Archtypes
- ftp files!
- scp files!
- SQL Share
- Myria
- Don’t even think about Sharepoint
- GitHub
- Heidi Sosik at WHOI: Flow cytometry images
- Jupyter
- What is the history of MODIS?
- Civilized?!
- GINA
- Ice2Ocean
- LiveOcean
- OOI…
- IOOS
- NOAA
- USGS cf Katherine Kuhn
- EPA STORET
- USDA
- NEON
- Ameriflux
- R2R (L-D)
- L-D Directories, GeoLink
- BCO DMO
- NSIDC
- Various approaches to ARGO
- Wild West?!
- JGI (Genomics)
- BLAST (as a tool) genes? proteins?
- CINERGI out of EarthCube (UCSD)
- GEOSS as a Meta Organization
- DOM at WHOI
- Geometabolimics Data Systems (GDS) Rob’s system for DOM
- GEOTRACES
- Harmonized world soil database
- FetchClimate???
- GEOLINK (authority tracking notification)
- David Shean’s greatest hits
- ESIP as a Meta Organization
- Scale
Triage
- The interview
- Who are you?
- What data problem are you trying to solve?
- What impact are you trying to have on your community?
- What are your use cases?
- Who are your producers?
- Your consumers?
- Your dependencies?
- Your data formats?
- Your data types?
- And so on
Old images